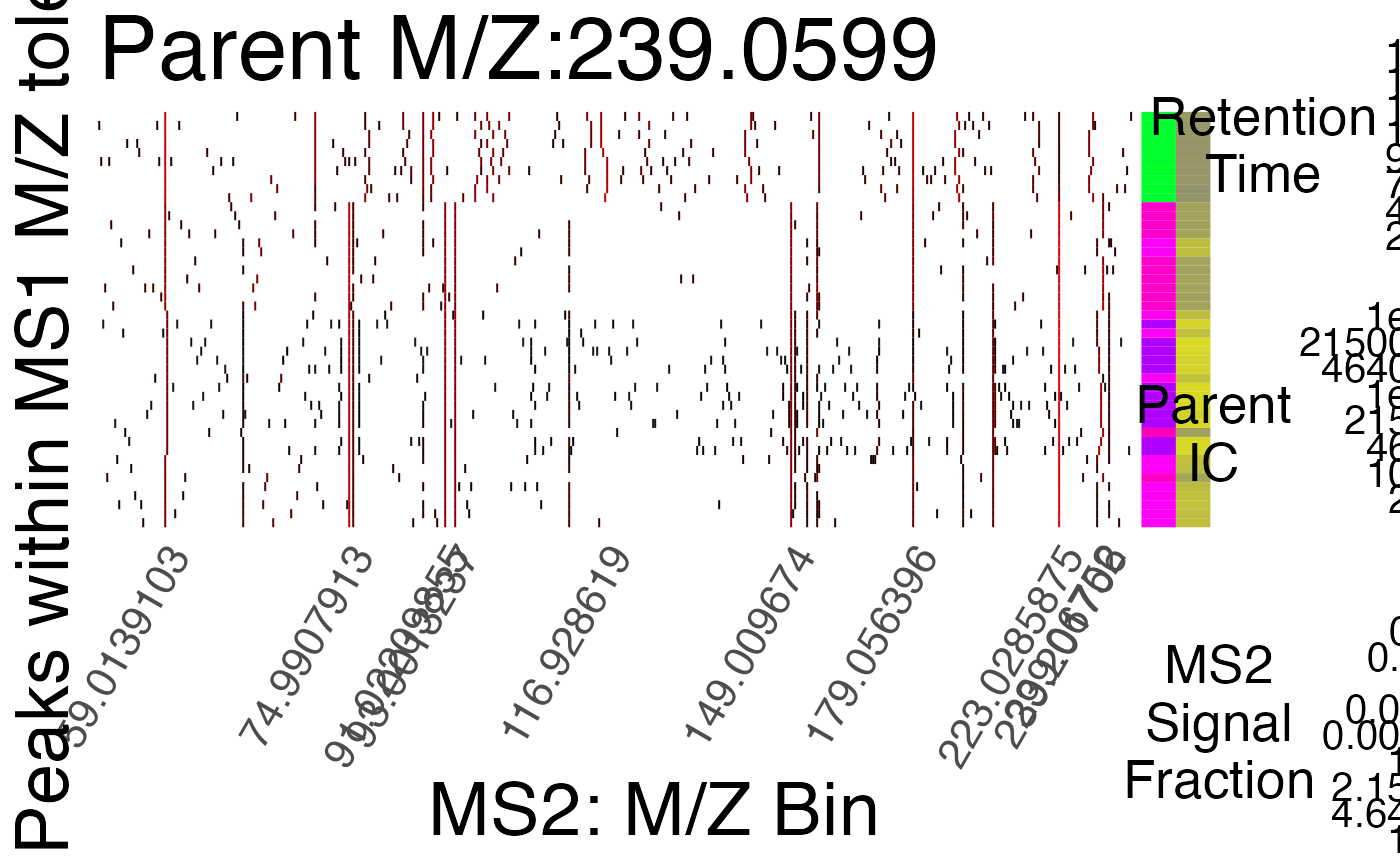
Find Consistent MS2 Fingerprints
find_consistent_ms2_fingerprints.RdCompare all MS2 fingerprints within the group to separate distinct groups based on MS2 profiles.
find_consistent_ms2_fingerprints(
mz_set,
MS2tol,
cosine_cutoff = 0.95,
create_plots = FALSE,
rt_range = NULL
)Arguments
- mz_set
a set of peaks with a consistent parent mass m/z which may contain multiple species with distinct RT / MS2 fingerprints.
- MS2tol
mass tolerance for grouping ms2 m/z values: see
build_clamr_config.- cosine_cutoff
Minimum cosine similarity between a pair of MS2 fragment profiles to group them into a common cluster.
- create_plots
TRUE/FALSE: should a summary plot be printed.
- rt_range
optional if create_plots is FALSE, if create_plots is TRUE, length two numeric vector specifying the lower and upper limits of retention time for an experiment.
Value
assignments of parent peaks to distinct ms2 peak groups
Examples
MS2tol <- build_clamr_config(list(MS2tol = "20ppm"))$MS2tol
find_consistent_ms2_fingerprints(
mz_set = clamr::mz_set_example,
MS2tol = MS2tol, create_plots = TRUE, rt_range = c(0, 22)
)
#> Warning: `select_()` was deprecated in dplyr 0.7.0.
#> Please use `select()` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.
#> Error in loadNamespace(x): there is no package called ‘fastcluster’